What Does Blue and Red Represent Multalin Alignment
Sequence Analysis Instructions
In order to predict your drug metabolizing phenotype from your CYP2D6 gene sequence, you must determine:
����������� 1)� If your PCR product even represents the human CYP2D6 gene,
����������� 2)� The location of your sequence within the CYP2D6 gene,
����������� 3)� Whether there are any sequencing errors or polymorphisms in your sequence,
and������ 4)� The effect(s) of any polymorphisms on CYP2D6 protein sequence.�
As you analyze your gene sequence, copy and paste your analyses and results into a text file for your final lab report.� If some factor, like the quality of your sequence, prevents you from carrying out the complete analysis, your grade will not be penalized�just complete as many of the steps below as possible, and include an explanation of why you could not complete the analysis in your final report.� Font appearing in bold green italics describes questions to answer and items to include in your final report.
Part 1:� Is your PCR product the human CYP2D6 gene?
1. Determine whether your gene sequence matches the published sequence for the human CYP2D6 gene.� To do this use GenBank, a nucleotide database run by the

Copy and paste your sequence into the search box, then click the button that says �BLAST!�

On the page that loads, click the �Format� button, and a new web page will appear.�

�
After a few moments, your blastn results will load in the new web page, showing which sequences in the NCBI database most closely match your sequence.
2. A graphic at the top of the blastn results page shows where each match aligns within your sequence.� The color of each match represents the alignment score, or the strength, of each match.�
 �
�
Below the graphic is a list of the database hits, including their scores and expectation values (E values).� The score of an alignment indicates how well your sequence aligns with a given sequence from the GenBank database, and takes into account such factors as gaps and mismatched bases.� The higher the score, the better the alignment.� E values indicate the significance of a match, and represent the expected number of random (chance) alignments that would have an equivalent or better scores than the one given for a particular hit.� Smaller E values correlate to higher alignment scores and thus indicate better matches.
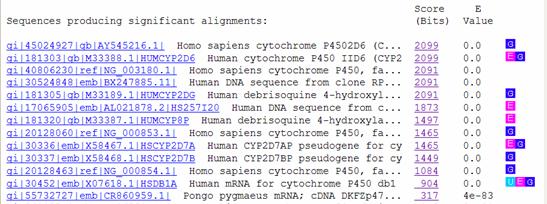
Below the list of hits, the sequence alignment for each match is presented.� This is where you can see how the two sequences align, including the location of any gaps or mismatched bases.� The alignment score and E value are also given here, along with a numerical summary of how many base matches and gaps there are in the alignment.

Include your interpretation of the blastn results in your lab report.� What are the highest 4 � 5 matches?� What are their relative E values?� Are they human genes?� Is human CYP2D6 the highest match?� If not, discuss possible reasons why not.
Note how BLAST provides a local alignment- only showing areas that it matched based upon the parameters. You can see small mismatches within the aligned sequence, but from the base numbers in the output, not all of your sequence may be shown. A program that provides a global alignment will try to make the best match over the whole sequence.
Part 2: Are there any sequencing errors or polymorphisms in your sequence?
- The genomic sequence of CYP2D6 is at
�http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=40806230&db=Nucleotide&dopt=GenBank.� Because the human genome sequence was obtained from an individual with the CYP2D6*5 allele (where the entire CYP2D6 gene is deleted), the wild-type CYP2D6 gene was sequenced separately.Scroll down the page until you reach the sequence of the gene.� Notice how each line has the base numbers on the left side.� When using a gene sequence for alignment or searching, these numbers in the middle of the sequence become problematic.� Therefore, to get rid of these numbers, there is a sequence display option called FASTA.� FASTA sequences lack numbers and line breaks.� For more information on FASTA format, see http://www.ebi.ac.uk/help/formats_frame.html and click or scroll down to the section on FASTA.

To display the sequence in FASTA format, look for the drop-down menu in the top left corner of the page�it is next to the word or button that says �Display.�� Change the display option from �GenBank� to �FASTA.�� If the page does not automatically re-load, click on the �Display� button (if present).

Copy and paste the FASTA version of the CYP2D6 genomic gene sequence (excluding the top line of text) into your report for easy access�you can delete it later.
- Several different websites will align sequences for you; one such program can be found at http://prodes.toulouse.inra.fr/multalin/multalin.html.
- In Aligning your PCR sequence with the genomic sequence of the CYP2D6 gene you will use a slightly modified version of the above genomic sequence (found at http://chemlife.umd.edu/classroom/bsci415/straney/lab/CYP2D6_genomic_coding.html) that removes the large portion of the gene upstream (5�) from the start codon�this will change the numbering of the bases to be consistent with allele nomenclature.� When you compare your sequence to known polymorphisms of the CYP2D6 gene, you will use a list of published polymorphisms found at http://www.imm.ki.se/CYPalleles/, and this site uses base numbering that begins at the start codon of the CYP2D6 gene.
Copy and paste the genomic sequence from the file into the white sequence box in Multalin.� Before the sequence, add a line that says �>genomic� to identify the sequence in the search results.�

Add a blank line below the sequence and add the PCR sequence with a separate line above it with a �>� and a title (such as �>rawPCR�)
Scroll down the page to �Optional Parameters.�� Under the heading �Alignment parameters,� find the drop-down box that says �Blosum62 - 12 - 2� (a default protein alignment algorithm) and change it to �DNA - 5 - 0� so that the program aligns a nucleotide sequence instead of an amino acid sequence. Also, change the �gap penalty at extremes� to both, it keeps mismatches near the ends from creating large gaps.

![]() Click the �Start MultAlin!� button to get your alignment.�
Click the �Start MultAlin!� button to get your alignment.�
� The results page will show the sequence alignment as a .gif image, which you can save in your report, or you can choose an option to display the results as an html page, from which you can copy and paste the results as colored text into your report�this feature is helpful because you can easily manipulate the size or font of the text to make it fit in your report, something you can�t do with a .gif file. If colors disappear upon cutting and pasting, you can instead save the html file to your disk, open it up in Word and then save it as a .doc file. Changing the font to a fixed point font (e.g. Courier) makes the line-to-line alignment match.�
� Another option on the page allows you to change the number of bases displayed per line (the �Maximum line length� default value is 130)�shrinking this value (60 is good) may help keep the alignment�s formatting in your report easier.
� The strength of alignment at each base is color-coded to help you quickly visualize differences between the sequences.� If you like you can change the colors the program uses to indicate different alignment consensus levels (the default options are black for no or neutral alignment, blue for low alignment, and red for high alignment).
� If you see matches of your PCR product in this alignment that break up your PCR product at the ends, stop and think if this makes sense. This would indicate that there is a large deletion in your allele, and an even greater rearrangement when you locate where the PCR primers are later. The alignment program can have problems matching the ends of the PCR product if it encounters mismatches (even with the end gap option), resulting in such a fragmented alignment. Test this by manually move any ends of the PCR sequence in the alignment to make a contiguous PCR sequence. Look at this alignment in the following analysis.
Copy and paste the alignment into your report. If color fonts disappear, save the html file, reopen it in Word, and then save it as a Word document. It is important to change the font to a �fixed� font, like Courier New and change the size of the font to retain the alignment�s original formatting.
- Carefully examine the sequence alignment for places where your sequence varies from the full-length genomic wild-type sequence (the CYP2D6*1 allele).� For each nucleotide difference, you will need to decide whether the discrepancy can be corrected (changed to the published sequence), whether it is an ignorable error because it occurs near the ends of your sequence (see below for more information on this error), or whether it is truly a polymorphic or heterozygous site.� Ultimately, after examining all the discrepancies between your sequence and the CYP2D6 gene sequence, you will make a corrected version of your allele sequence(s) for use in later analysis steps.�
To determine which type of the above differences you have, you will need to locate each discrepancy on your sequence�s chromatogram�this is the raw sequence data obtained by the sequencing machine.� To open the chromatogram file, you will need to download a free software program from http://www.technelysium.com.au/chromas.html (or Editview for Mac at http://www.appliedbiosystems.com/support/software/dnaseq/installs.cfm).� The sequencing facility uses a similar base-calling program that reads and interprets the peaks in your chromatogram to give the location and identity of each base in your sequence. Use the chromatogram as your source to look for evidence that you sequence is different than wild-type.
�
Types of discrepancies you may encounter in your sequence:
� Your sequence differs from the wild-type sequence at one base. � Examine your chromatogram�sometimes the base-calling program makes errors that you can correct by eye.� When examining your chromatogram, you will see peaks in your chromatogram that are not consistent with the base(s) that the program interprets.


If you should find similar errors in your chromatogram, go with what the peaks imply should be the sequence and change your sequence (if necessary) to reflect what the peaks show.� Peaks that differ from the published wild-type CYP2D6 gene sequence may represent single nucleotide polymorphisms (SNPs)�highlight or change the color of the base to make it stand out.
� 
Your sequence contains an �N� base. � Ns occur when the base-calling program cannot resolve the identity of the base.� You may be able to correct the N by eye after examining the chromatogram.�
However, if you cannot easily identify the base, evaluate if it is still consistent with the wild-type sequence, given the uncertainty. If it is inconsistent with wild type, leave it as an N.�


One complicating factor is heterozygosity�you may have two different alleles that differ by one base at this location.� This will appear as two overlapping peaks in the chromatogram.� One of the peaks may represent an allele with the wild-type (normal genomic) sequence, and the other peak may represent an allele with a SNP at that location.
If you have a heterozygous base, leave the N in the sequence and mark it with a different color or highlight than you are using for your polymorphic sites.
� Either your sequence or the genomic sequence has a gap (denoted by - ).� Gaps can result when there has been an insertion or a deletion in one of the sequences.� If your sequence has the gap, there was a deletion in your sequence; conversely, if the genomic sequence has the gap, then there was an insertion in your sequence.� Examine your chromatogram�sometimes the base-calling program can miss a base, especially when there is a string of the same nucleotide (like AAAA).� The following two chromatograms show portions of a sequence with runs of C residues in which the base-calling program has misidentified the number of C bases.� In the chromatogram on the left, only three C bases were identified by the program, but there are four blue peaks; here, the program omitted a C base.� In the chromatogram on the right, the program has identified four C bases, when there are only three blue peaks; here, the program added an extra C base.

�
Look at the spacing and width of peaks in relation to other nearby peaks; you may or may not be able to correct the gap.� If you cannot correct the gap, leave it in the sequence and mark it with a new color or highlight�there are known polymorphisms of CYP2D6 that have small deletions or insertions.
� You seem to have many mismatches in close proximity to each other, especially near the 5� or 3� ends of your sequence. � The first 10-30 and last 100-250 bases of sequence from the machine can be unreliable and full of uncorrectable errors. Peak broadening makes counting the number of bases in a string of the same base difficult; as a result, we will likely only get about 800 bases of reliable sequence from our PCR product. Again, focus upon looking for clear evidence in the chromatogram that there is a difference from the wild-type sequence, given this uncertainty. Use your judgment to determine where this becomes too unreliable and �chop off� the ends from your final corrected sequence.
After examining the discrepancies between your sequence and the wild-type CYP2D6 genomic sequence, create a corrected version of your sequence in FASTA format that incorporates any changes you have made, removes the ends of your sequence up to where reliable sequence begins, and removes any gap symbols in your sequence (these will be added back in by an alignment program later).� In positions where you believe there are two possible nucleotides (like at a heterozygous site), make two versions of your sequence, one to represent each allele.� Include your corrected sequence(s) in your final report, along with a brief description of the changes you made and reasons for doing so.�
You may wish to run a blastn search with your corrected sequence to make sure that you did not �over-correct� your sequence.� Make sure that the top hits are to the human CYP2D6 gene.
Part 3:� Determine the context of your PCR sequence within the CYP2D6 gene
1. Pull up the mRNA sequence of CYP2D6 at http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=NM_000106.4.� Change the display option of this page to FASTA format, as above, to give the mRNA sequence as solid text without numbers or line breaks.� Copy the FASTA version of the mRNA sequence into your report. �
In your report, make note of the coding region (CDS) of the mRNA sequence by changing the font color or by bolding the first start codon (ATG at position 91-93) and stop codon (AAT at position 1585-1587). Highlight the 5� untranslated region before the first ATG.
2. Trim the first 90 bases before the first ATG off your mRNA sequence; this will make the numbering in the alignment consistent for the polymorphisms. Align your sequence with the genomic and mRNA CYP2D6 sequences to see where your PCR product�s sequence is located within the CYP2D6 gene and whether any introns lie in the sequence. The Multalin program you used above ( http://prodes.toulouse.inra.fr/multalin/multalin.html ) can do multiple sequence alignments. Remember to choose the DNA alignment parameters, the end gap option and use the shorter version of CYP2D6 genomic DNA (beginning at the ATG start codon at http://chemlife.umd.edu/classroom/bsci415/straney/lab/CYP2D6_genomic_coding.html ), since the database version is be too large for a three-way comparison.
Copy and paste the genomic sequence from your report into the white sequence box.� Before the sequence, add a line that says �>genomic� to identify the sequence in the search results.�

After the genomic sequence, add an empty line to denote the end of the sequence, then insert the mRNA sequence with an identifying line before it, like �>mRNA�. Use only the portion of the mRNA sequence after the first ATG (do not paste in the highlighted 5�untranslated region). This will keep the numbering consistent with allele SNP positions in the literature.
After the genomic sequence, add an empty line to denote the end of the sequence, then insert your corrected PCR sequence with an identifier of �>correctedPCR�
� Note that the poly(A) tail on the 3� end of the mRNA sequence will not align with the genomic sequence.
Paste this alignment into your report Again, if color fonts disappear, save the html file, reopen it in Word, and then save it as a Word document. It is important to change the font to a �fixed� font, like Courier New and change the size of the font to retain the alignment�s original formatting. Changing the number of characters per line also helps formatting. If you wish, you can delete the raw, unaligned genomic and mRNA sequences from your report and just include the alignment; however, make sure the start and stop codons are clearly marked in your alignment.
How many exons are there in the CYP2D6 gene?
Which exon(s) and/or intron(s) fall in your sequence?
Discuss how your sequence varies from the wild-type sequence. Do the differences occur in exons or introns? Note that changes in the first 2 bases at the beginning and end of an intron can eliminate splice sites, creating different splicing.
- Now find the location of the PCR and sequencing primers we used to amplify this gene in the sequence alignment.� Their sequences are:
����������� (PCR)� Ex6F2:��������� 5�� AAGAAGTCGCTGGAGCAGTGGGTGA����� 3�
����������������������� Ex11R:��������� 5�� ACCGATGACAGGTTGGTGATGAGTGT���� 3�
����������������������� 4Flong:�������� ��� 5�� GCCTTTGTGCCGCCTTCGCCAACCACT��� 3�
����������������������� 2Rlong:����������� 5�� CCCTCGGCCCCTGCACTGTTTCCCAGAT�� 3�
�����������
����������� (Sequencing)��� ���������� 5�� TGATGGGCAGAAGGGCACAAA� 3�
Note that the Ex11R and 2Rlong primers are in the reverse direction (3� � 5� when read from left to right), so you will need to find their reverse complements (5� � 3� from left to right), either by hand or by using a program like http://bioinformatics.org/sms/rev-comp.html (and using the �reverse-complement� option).
To find the PCR primers in your alignment, you can search for them by hand or use a search function in your text program (e.g.: Find in Word). Note the spaces in the sequence may make find difficult- you can remove all spaces with replace.� The area between the PCR primers is the region that was amplified in the PCR reaction, and the sequence downstream (3�) of the sequencing primer is the portion of the PCR product that was sequenced by the sequencing facility.
Highlight or change the font color to make the primers stand out, and include an identification key that details these changes in your final report.
� How far downstream of the sequencing and PCR primers does your sequence start?
� Are they in introns or exons?
3.� Compare your corrected sequence(s) to the known polymorphisms in the CYP2D6 gene.� The numbering of polymorphisms in databases should be applicable to your sequence comparison as long as the genomic/mRNA sequence starting at ATG was used.
�One useful list is on http://www.imm.ki.se/CYPalleles/ .� This database includes the enzyme activities of the alleles. See the sequence alignment program below for a quick graphic comparison.
To make your life easier, the FASTA-style sequences of selected CYP2D6 alleles (instead of a list of the polymorphisms) have been compiled at http://chemlife.umd.edu/classroom/bsci415/straney/lab/CYP2D6allelesDNA.html.� The polymorphisms in each allele appear in a different color from the rest of the sequence.�
� Use the sequence alignment program at http://prodes.toulouse.inra.fr/multalin/multalin.html to align your corrected sequence(s) (in FASTA format) with the various CYP2D6 alleles (just copy and paste the CYP2D6 allele sequences from the above link, then add your sequence(s)).� Your PCR product is shorter than the allele sequences which extend from primer 4Flong to 2Rlong. To make a meaningful tree, pad your sequence with wild-type sequence out to the 4Flong and 2Rlong primers from your alignment.
Copy this alignment into your final report, along with a discussion of the differences you see between your sequence and the various CYP2D6 alleles.� Does your sequence match any known CYP2D6 alleles?� If so, what is your predicted drug metabolizing phenotype (ex: does your sequence match an allele with a poor metabolizing phenotype)?� A good reference for the range of activities for different alleles is figure 1 in the paper: http://www.aapsj.org/articles/ps0204/ps020433/ps020433.pdf . Also, look at activities of known alleles at http://www.imm.ki.se/CYPalleles/ if yours matches a known allele.� Do you have two different alleles, is there evidence for heterozygosity (do each of your predicted alleles match known CYP2D6 alleles)?
����������� If you can not find your specific polymorphisms in the CYP alleles database, check the human SNP database at http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=1565&chooseRs=coding
(you can get here by searching under NCBI SNP for CYP2D6). Go to the first section saying �Gene Model (mRNA..)�.Click on radio button for in �gene region� and hit the �refresh� button. This will show SNPs in introns as well. The numbering of polymorphisms is based upon the DNA contig. The ATG starts at position 27977309 and goes down, since the gene is on the bottom strand. Is your SNP in this database? It is organized by SNP alone and does not include grouping of SNPs as alleles.
- The MultAlin program also has a feature that will create a phylogenetic evolutionary tree that shows the relatedness of the aligned sequences.� To make this tree, click on the �Alignment and tree description (rtd)� link that appears below the alignment results.�

Click on the small tree graphic in the white box that appears above the sequences you input.� The tree will appear in a new window; copy and paste this tree into your report.

Is your sequence closely related to a known allele, or is it distinct enough to create a new subclass?
Part 4:� Determine the effects of your sequence�s polymorphisms on your protein sequence
- Obtain the coding portion of your corrected sequence.� To determine which part of your sequence is coding sequence, you can either use your alignment from part 2, when you aligned the genomic and mRNA sequences of CYP2D6 with your unmodified sequence, or you can run a new alignment using your corrected sequence(s) in place of your unmodified sequence.� Then look for areas in the alignment where all three sequences align (with default settings, these are indicated by red font)�these portions of the sequence are the coding portions.� �Splice� your corrected sequence by cutting and pasting together the exonic (coding) portions of your sequence, and deleting the intronic (noncoding) portions.� Put your sequence into FASTA format.�
- Using this same splicing technique, obtain the coding portion of the CYP2D6 gene that corresponds to (matches) your sequence.
- Translate this coding portion of the wild-type CYP2D6 gene by using the ExPASy translation tool found at http://us.expasy.org/tools/dna.html.� Select the output format �Compact (�M�, �-�, no spaces).��

Your results will come back with six different reading frames�choose the frame in the 5� � 3� direction that does not have any �-� symbols (these stand for stop codons), since the coding portion of a gene will not have stop codons (except for one at the 3� end). If the correct reading frame is not clear, use the CYP2D6*1 protein sequence from: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&qty=1&c_start=1&list_uids=NP_000097.2&uids=&dopt=fasta&dispmax=5&sendto=&from=begin&to=end&extrafeatpresent=1&ef_CDD=8&ef_MGC=16&ef_HPRD=32&ef_STS=64&ef_tRNA=128&ef_microRNA=256&ef_Exon=512

- Using the ExPASy translate tool, translate the coding portion of your corrected sequence.� Select the translation product with the same reading frame that gave you no stop codons with the coding portion of the CYP2D6 gene (with the above example, you would select the �5�3� Frame 2� reading frame for your sequence).���
- To detect differences between your translation product and that of the wild-type CYP2D6 gene, run an alignment of the two sequences with the program at http://prodes.toulouse.inra.fr/multalin/multalin.html, using the default �Blosum62 - 12 - 2� setting for the Symbol comparison Table instead of �DNA - 5 - 0.��
Include the translation products of both the gene sequence and your corrected sequence(s) and discuss any differences you see between the two.
� If you had any polymorphisms, did they affect the translation product or were they silent mutations, were they in an exon or in an intron?
� If you had amino acid substitutions, were they conservative or nonconservative?� How might the substitution(s) affect the function of your gene?
� If you have a single or double base pair insertion or deletion, these will alter the reading frame of your protein, and the part of your sequence downstream from this insertion/deletion will not align with the wild-type protein sequence.� Do you have evidence of this in your sequence?
![]()
��������������������������������������������������� �������������������������������� Mollie Minear, October 2005
arledgespearknigh91.blogspot.com
Source: https://science.umd.edu/classroom/bsci348g/straney/lab/PCR%20analysis.html
0 Response to "What Does Blue and Red Represent Multalin Alignment"
Post a Comment